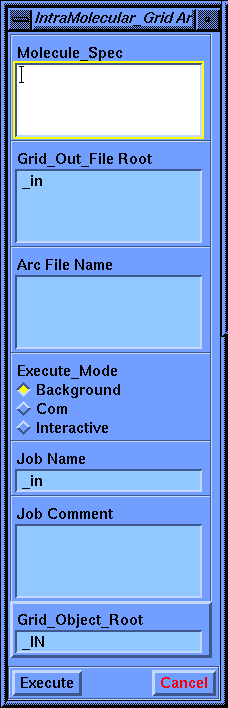

Background automatically spawns a background job to execute the calculation. When the job is completed a message is printed in the information area.
Com writes the background input file but does not initiate the calculation. You may submit the job manually at a later time.
Interactive performs the calculation in immediate mode. Note that this locks out InsightII for other functions until the calculation is complete. The calculation time for Hint grids and tables varies from a few seconds to several hours depending on complexity.